Neal Woodbury
Professor: Department of Chemistry and Biochemistry,
Arizona State University
|
 |
One of the most important and fundamental
subjects in biochemistry is the molecular level control of reaction
mechanism by protein structure. The protein is the ultimate hi-tech
solvent in which to run a reaction. It provides complete three-dimensional
control over polarity, group reactivity, and steric constrains.
In addition, once a biochemical reaction is initiated, a preprogrammed
sequence of changes in these parameters occurs which can guide
chemical reactions along very specific mechanistic pathways.
Most of the pictures on this page are links to articles from our lab. Many more articles are provided with links under "Publications" below.
Our major interest is in the study of molecular dynamics and
mechanism in protein mediated chemical reactions. The difficulty
one usually faces in such research is that reaction rates of most
enzymes are controlled by diffusion. Because of this, it is very
difficult to observe reaction intermediates that are formed and
decay on time scales faster than bulk diffusion and thus the study
of enzyme mechanism is normally by indirect methods. Usually,
intermediates must be trapped by altering the structure of the
reactants or adding reagents which block the reaction at a specific
step.
In order to overcome these problems,
we have chosen to study systems in which the reactions are initiated
by light. Modern laser technology allows one to generate pulses
of light only 10's of femtoseconds long (a femtosecond is one
one thousandth of a trillionth of a second) allowing one extraordinary
kinetic control of the reaction. By observing the changes in the
optical properties of the system (absorption and fluorescence)
as a function of time after the reaction is initiated, changes
in the electronic states of the system can be followed directly,
and mechanism can be inferred from the kinetic, thermodynamic
and spectral properties of the intermediates observed.
One of the systems we study is the photosynthetic
reaction center. This is the protein-cofactor complex that performs
the primary reactions of solar energy conversion in plants and
photosynthetic bacteria. These are extremely important reactions
as all of the biological energy sources on the earth, including
all fossil fuels, result from light energy absorbed and converted
by this process. The initial events of photosynthesis
are a series of electron transfer reactions between neighboring
cofactors embedded in a protein complex. The result of these reactions
is a charge separation across a cellular membrane: a biological
solar powered battery. The high energy charge separated state
thus formed can be used to power the translocation of protons
across the membrane and this in turn powers the production of
ATP, the energy currency of a living cell.
There is no diffusion involved in photosynthetic
electron transfer, and thus the reaction can be initiated solely
by light absorption. The reaction center cofactors have absorption
and emission spectra in the ultraviolet, visible and near infrared
regions of the spectrum, and are thus easily monitored on ultrafast
time scales. It is possible in this way to literally watch the
electron hop from cofactor to cofactor in the complex, monitoring
the spectral, kinetic and thermodynamic properties of the intermediates
formed along the way. This results in tremendously detailed information
about the reaction mechanism.
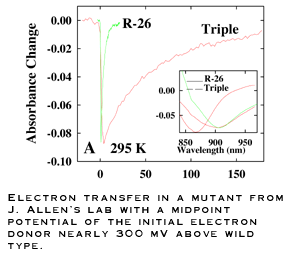
The reaction center complexes we study are derived from bacterial
rather than plant sources. Though the structure and function of
both the prokaryotic and eukaryotic reaction centers of this type
are thought to be very similar, the bacterial system offers many
biochemical advantages. The two most critical advantages are the
availability of a three-dimensional structure of the complex and
the ability to easily modify the protein structure through recombinant
DNA techniques. We have been very fortunate to collaborate closely
on much of our work with Professor James
Allen's laboratory. Collaboratively we perform directed mutagenesis,
expression and isolation of altered reaction centers. These mutant
reaction centers are then subjected to spectroscopic analysis,
as described above, as well a structural analysis by X-ray crystallography
in Dr. Allen's laboratory. By judiciously selecting the amino
acid residues to alter and then carefully evaluating the effects
of those changes on reaction mechanism, we are beginning to learn
the important rules which govern the static and dynamic relationships
between protein structure and chemical mechanism.
Structural work has suggested that the
chromophores in the antenna form a closely packed ring. Just what
are the true optical properties of this ring? Does it behave like
many independent molecules or like one giant molecular complex
or somewhere in-between? How does this effect the energy transfer
to the reaction center in the center of this ring? Why is the
energy transfer process as temperature independent as it apparently
is? How does the energy transfer process work with regards to
the thermodynamics of the charge separated state in the reaction
center?
We are also exploring other biochemical systems that do not
normally contain natural optical probes by gentically engineering
such probes into them. This is of particular interest in light
of the recent advances in single molecule spectroscopy. While
other biochemical systems may not have reactions that are initiated
by light, if they have an optical reporter group engineered into
them, their stepwise mechanisms may still available for spectroscopic
investigation in real time if we look at one molecule at a time.
This leads to the possibility of single molecular probes for the
elucidation of binding interactions, protein dynamics and multistep
mechanisms. At the moment, we have built
DNA-sequence specific polypeptide probes with optical reporter
groups attached whose fluorescence properties are extremely
sensitive to their interaction with DNA and even to what sequence
of DNA they interact with. We have also constructed and are performing
measurements on a confocal microscope based
single molecule spectrophotometer. This instrument has been
used to investigate the
binding properties of the DNA protein system described above at
the single molecule level.
Finally, we have intiated work in the area of directed evolution. Specifically, we are building high-throughput systems for both viewing large numbers of bacterial colonies expressing a library of modified proteins and we are developing techniques for optically selecting individual colonies (using imaged light to kill the colonies that we do not want at each round in the directed evolution procedure). At the moment, the focus of this research is the merge a binding site for glucose into the green fluorescent protein structure and then introduce variation into this fusion and select for fluorescence coupled to glucose binding.
One of the most attractive aspects of our work is its interdisciplinary
nature. Students and postdocs in our laboratory will very likely
be involved in problems of protein genetic engineering and expression,
biochemical analysis and purification of protein complexes, and
detailed evaluation of reaction mechanism by performing and interpreting
ultrafast spectroscopic measurements. Mastering several rather
different disciplines is a challenging goal, but ultimately provides
a researcher with a broad-based understanding of biochemistry
and biophysics which can be applied to many different problems.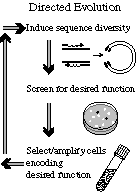
I participate a number of different training program at the undergraduate, graduate and postdoctoral level. Students and postdocs interested in working with me may wish to consult the web pages for the Research Education for Undergraduates program at ASU (undergraduate summer support), the Graduate Research Training Program in Energy Transduction (graduate student support), or the Research Training Progam in Optical Biomolecular Devices (graduate and postdoctoral support). Those interested in our work should also feel free to contact me directly for reprints or other information.
Return to Woodbury's Home Page
Neal Woodbury
Department of Chemistry & Biochemistry
Arizona State University
Box 871604
Tempe, AZ 85287-1604 USA
Tel: 1-(480) 965-3294
Fax: 1-(480) 965-2747
Email: nwoodbury@asu.edu
Office Room Number: Physical Sciences PS D-105
Lab Room Number: Physical Sciences PS D-114