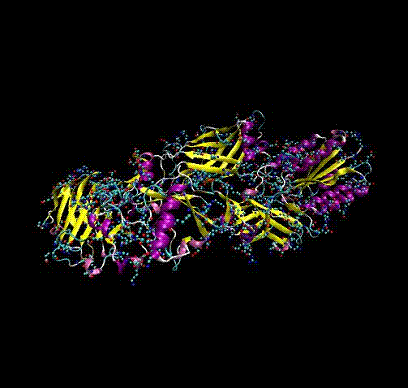
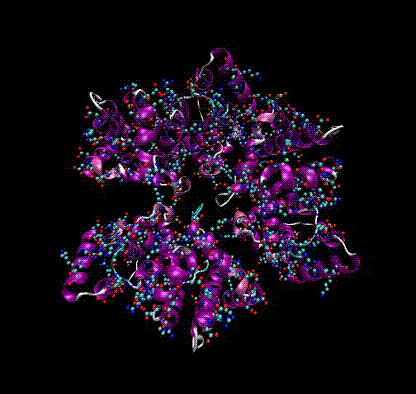
Biophysics
I am interested in how proteins fold (aren’t we all!), and how to determine their structure from minimal diffraction data. (Collaboration with Prof J.C.H. Spence + group and Prof D.K. Saldin, University of Wisconsin, Milwaukee). At present I am investigating whether one can use non-linear refinement techniques to directly derive Ramachandran angles (that define the polypeptide chain) from diffraction data.
Reference
”Dose, exposure time and resolution in serial X-ray crystallography”, D. Starodub, P. Rez, G. Hembree, M. Howells, D. Shapiro, H.N. Chapman, P. Fromme, K. Schmidt, U. Weierstall, R.B. Doak and J.C.H. Spence, J. Synchrotron Rad., (2008) 15, 62-73.
Electromagnetic Response of Large Biomolecules
Funded by Defense Threat Reduction Agency
Graduate students Adam Mott, Ani Kadne, postdoc Saravana Thirumuruganandham
Can any part of the electromagnetic spectrum be used to distinguish biothreats (anthrax, smallpox) from innocuous biomolecules?
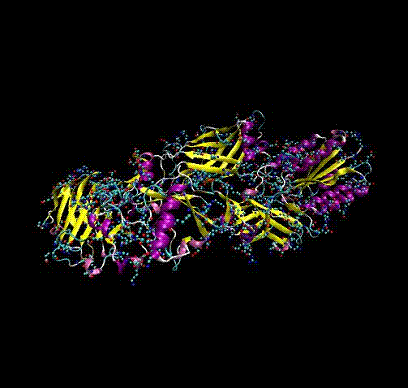
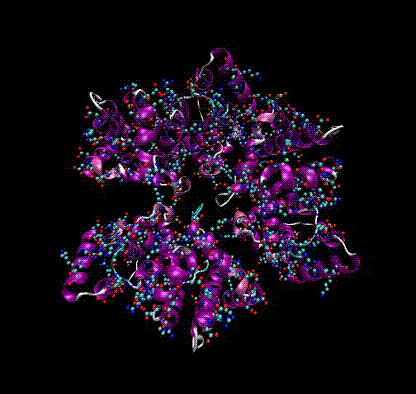
We are calculating the electromagnetic response of large biomolecules from first principles from THz frequencies through the infra-red and optical range. This involves diagonalizing some very large matrices calculated using force fields from molecular dynamics codes such as CHARMM.
 oooooooo
oooooooo

oooooo
 oooooooo
oooooooo

Fortunately they are sparse (have a lot of zeroes) and we are developing innovative techniques for matrix diagonalization using large parallel clusters (such as our local Saguarro cluster). We have started by checking our methods on small molecules and comparing the results with quantum chemistry calculations
 oooo
oooo