Group Members |
|
From Left: Raimund Fromme, Alan Lee, Srinivas Makam, Jane Gong, Petra Fromme, Katerina Dörner,
Chris Kupitz, Brenda Reeder, Kim Rendek, Ingo Grotjohann, Justin Flory, Tina Esquerra, James Zook
Danielle Cobb, Mae Pittman, Robin Paul, Jose Garcia |
Faculty Research Associates and Post Doctoral Researchers |
- Dr. Katerina Dörner

|
Katerina is part of the Center for Membrane Proteins in Infectious Diseases (MPID). She works on protein purification, biophysical characterization and crystallization. She is interested in the structure and function of the small envelope (E) protein from transmissible gastroenteritis coronavirus as well as in membrane proteins from the bacterial human pathogen Francisella tularensis. Structural information of these proteins are fundamental to understand virulence of these organisms and therefore essential for vaccine development. Based on E. coli as an expression host, Katerina develops and optimizes purification protocols to isolate the heterologous produced target proteins. Purified proteins are characterized by Dynamic light scattering (DLS) and Circular dichroism (CD) spectroscopy to optimize crystallization behavior as a prerequisite for X-ray structure analysis.
|
|
|
The inner sanctum of Photosynthesis the structure and function of Photosystem I and II is since my master thesis and PhD the fundament of my research interest. Starting in 2004 I have the opportunity to work on crystal structures of various proteins in the broad field of photosynthesis. The membrane proteins are the most interesting and challenging proteins at all. Currently the protein data bank has above 80,000 structures, in contrast the number of known unique membrane protein structures is still around 300 . Therefore the most important proteins are in their majority still unknown by their structure, beside the fact that the numeric share is at least one third of all proteins. The field of membrane protein structure determination is still in the beginning with a clear growing impact to many research topics in chemistry, biochemistry and biology. Diffraction with the X-ray femtosecond Free Electron Laser (FEL) is the newest tool to provide dynamic information about structures. See Chapman et al. (2011) Nature 470, 73–77. Boutet et al. (2012) Sciencexpress from 31 May 2012.
|
- Dr. Jose Martin Garcia

|
Jose is a Postdoctoral Researcher Associate at Arizona State University. He is working on the purification, biophysical characterization (Size Exclusion Chromatography, Dynamic light scattering, Circular dichroism and Transmission Electron Microscopy), crystallization and structure determination of membrane proteins in the Center for Membrane Proteins in Infectious Diseases (MPID Center). Currently he is working on various targets from Francisella Tularensis organism.
|
|
|
No information regarding Dr. Grotjohann is available at this time.
|
- Kim Rendek

Kim Rendek is currently a Postdoctoral Research Associate at Arizona State University focusing mainly on bioenergy, biochemistry, molecular biology, and X-ray crystallography. Her primary work includes the design of an artificial oxygen-evolving complex (aOEC) that will be incorporated into solar fuel cells as part of a U.S. DOE-sponsored Energy Frontier Research Center (EFRC) project, and also the crystallization of Photosystem II from cyanobacteria. The need for a renewable and sustainable energy source is crucial, and this work adopts a bioinspired approach, emulating the water-slitting capabilities of Photosystem II, nature’s best kept secret. In cyanobacteria and plants, due to the instability of Photosystem II’s primary electron donor, the highly oxidizing P680+, Photosystem II’s core protein, D1, is degraded every half hour in the presence of sunlight. Thus, it is necessary for the aOEC to be contained in a stable framework, without the presence of P680+. The stable framework used for this project is a three-dimensional DNA tetrahedron, which is used to encapsulate synthetic peptide sequences based on the high-resolution X-ray structure of cyanobacterial Photosystem II, which reveals the amino acids that provide ligations to the manganese cluster (4MnCaCl).
|
As a preliminary study, the DNA tetrahedron itself has been characterized using X-ray crystallography, which will provide precise atomic positions that will be used to directly attach synthetic peptide sequences. Nanocrystals of the DNA tetrahedron have been obtained and analyzed using the free-electron laser at the LCLS at Stanford University. This state-of-the-art technique would allow for bypassing of the radiation damage process often encountered in nucleotide crystallography.
The overall goal is to successfully design, assemble, and characterize the aOEC through the use of X-ray crystallography, electrochemistry, and electron paramagnetic resonance (EPR). As a proof of concept, metal-containing porphyrins, specifically Fe(III) meso-tetra(4-sulfonatophenyl) porphyrin chloride, have been implemented for testing of the encapsulation of a metal-containing molecule inside of the DNA cage. Upon alteration of the spin state of the metal through coordination of two orthogonally oriented peptides covalently attached to the DNA tetrahedron which contain terminal histidine residues, iron (III) switches from its EPR-active state (high-spin) to low-spin and the assembly can be analyzed electrochemically. Upon assembly of the complete metal-containing center (both porphyrin molecule and aOEC), x-ray crystallography, EPR, and electrochemistry have been used to verify functionality in the stable DNA framework.
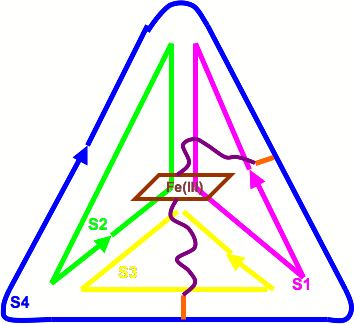
|
- Dr. James Zook

James Zook is a 2007 graduate from the University of Wisconsin - Eau Claire with a B.S. in Biochemistry and Molecular Biology. He joined the Fromme lab in 2007 as a graduate student and completed his PhD in 2012.
His graduate research centered around the structural characterization of membrane proteins using high-resolution nuclear magnetic resonance, circular dichroim, and light scattering with an emphasis on high resolution structure of membrane proteins using NMR.
|
James is part of the Center for Membrane Proteins in Infectious Diseases (MPID). He works on protein purification, biophysical characterization and crystallization. He is interested in the structure and function of proteins involved with the pathogenicity of Francisella tularensis, the bacteria responsible for the disease tularemia (commonly known as rabbit fever).
His current target is a protein that may share some similarities with the Yersinia Autotransporter Protein (Yap). These proteins are, in part, what made Yersinia Pestis so devestating in Europe during the Black Death which killed over 1/3 of the population in the mid 1300's.
Understanding how Yaps function in the role of pathogenesis is crucial to understanding why tularemia is so infective and can potentially help parts of the world where tularemia is a persistant problem.
|
Ph.D. Students |
- Shibom Basu

|
Shibom is a graduate student from India, working on data analysis part of Femtosecond Nano-Crystallography project, mostly focused on solving x-ray structure along with analyzing raw crystallographic data.
- Justin Flory

Justin Flory is a 2002 graduate from the Sonoma State University with a B.S. in physics. He is currently in the Biological Design program here at ASU.
Justin is deeply concerned about climate change and the impact of human development on the environment. Human society depends on the environment for many services like energy, clean air and water. Justin is a part of the Center for Bio-Inspired Solar Fuel Production sponsored by the Department of Energy, which is developing low cost methods to convert solar energy into hydrogen gas as a replacement for fossil fuels.
|
Justin is focused is on designing a catalyst inspired by Photosystem II (PSII) to oxidize water as a sustainable source of electrons in a solar fuel cell. However, PSII is damaged under full sunlight with a half life of 30 minutes and must be reassembled. His strategy is to synthesize the peptides surrounding the catalytic active site of PSII and reassemble the active site inside a stable scaffold made of DNA folded into a three dimensional nanostructure.
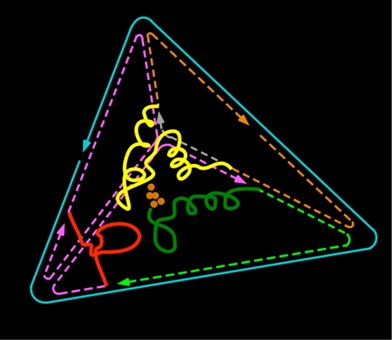
Read more about Justin Flory by visiting his ASU Directory Profile.
- Zhen Gong

Zhen Gong is a 2008 graduate from China Pharmaceutical University with a B.S. in Traditional Chinese Pharmacy. She joined the Fromme lab in 2009.
Zhen’s research is on the structure determination of the recombinant truncated HIV-1 gp41 consisting of the Membrane Proximal and Transmembrane Domains. The membrane proximal region (MPR) of gp41 is highly conserved across various clades and is the target of broadly neutralizing and transcytosis blocking monoclonal antibodies 2F5 and 4E10, but its native thee dimensional structure is not known. The generally accepted topology model for gp41 predicts that it has a single membrane-spanning domain with its C-terminus in the interior.
|
To facilitate its structural determination, we expressed in Escherichia coli a truncated form of gp41 consisting of the MPR and transmembrane (TM) domains (MPR-TM), fused to Mistic, a Bacillus subtilis integral membrane protein that was previously shown to promote the accumulation in bacteria of high levels of correctly folded membrane proteins. The His-tagged recombinant protein was extracted by detergent and purified by metal affinity chromatography. The Mistic moiety was then removed by Tobacco Etch Virus (TEV) protease cleavage. Final purification of the MPR-TM was accomplished using anion exchange column chromatography in the presence of detergent. The purified MPR-TM was determined by SDS-PAGE and MALDI. The Circular Dichroism data of MPR-TM show that it is alpha helix. The Dynamic light scattering data indicate that MPR-TM is monodisperse, which means it is good for crystallization. Thousands of different conditions have been tested for crystallization. Some conditions are promising but furthur improvements are still needed.
|
- Christopher Kupitz

(Photo: SLAC National Accelerator Laboratory)
Christopher Kupitz is a 2008 graduate from the Michigan Technological University with a B.S. in Chemistry. He joined the Fromme lab in 2010 as a graduate student.
|
Christopher's research is focused on growth of membrane proteins for use in and method development of a new technique, Femtosecond Serial Nanocrystallography. Specifically working with photosystem proteins, Photosystem I and II, and co-crystallizing Photosystem I and ferredoxin. Using the world's first high energy free electron laser at LCLS in Stanford, we are developing a technique where we can image the movement of the proteins in their various excited states by first exciting them with a laser, followed by a powerful x-ray pulse to collect the diffraction pattern. In this way we hope to be able to create a "molecular movie" of the conformational changes that occur during the water splitting/oxygen evolution process. Unravelling the mechanism of water splitting by PSII will allow us to develop better methods for bioenergy through the creation of hydrogen as a fuel source.
|
- Alan Lee

|
Alan Lee's research is on the structure determination of fusion protein consist of cholera toxin B-subunit (CTB) and membrane proximal region (MPR) of gp41 in the human immunodeciciency virus (HIV). CTB-MPR is a candidate AIDS vaccine that has shown to elicit HIV-transcytosis blocking antibodies. To further improve the design of the vaccine, it is important to determine the three-dimensional structure of the CTB-MPR.
|
- Robin Paul

My project constitutes studying complementary chromatic adaptation (CCA) in a newly discovered cyanobacterium Leptolyngbya Heron Island (L.HI) isolated from Heron Island, Australia. CCA response constitutes the change in expression of phycobiliproteins, where there is change in the Phycoerythrin/Phycocyanin expression ratio with change in light conditions from red to green light. However, this species of cyanobacteria shows a class II CCA response i.e it only changes Phycoerythrin with change in light conditions leaving Phycocyanin mostly constant. This brings about unequal light capture between the two light conditions. We intend to study how this affects the electron transfer chain in photosynthesis. Our experimental results suggest that this in turn affects the Photosystem I/Photosystem II (PS I/PS II) ratio. We have found that the PS I/PS II ratio is higher in red light adapted cells than in green light adapted cells. This may be an adaptation to balance overall electron flow in the photosynthetic electron transfer chain.
|
However, to completely understand this phenomenon we need to characterize all the regulatory genes involved in the expression of the phycobiliproteins and photosystems. For this, we are currently sequencing the genome of this cyanobacterium. This is being done using the next-generation Ion-torrent sequencer installed in School of Life Sciences, ASU. In the future we hope to identify genes (or the lack of certain genes) which are responsible for the class II CCA response and the change in PS I/PS II ratio with change in light conditions.
|
- Jayhow Yang

JayHow Yang is a third year Ph.D. student in Biochemistry at Arizona State University, joining the Fromme Research Group in January 2009. Prior to beginning his graduate studies at ASU, in July 2001, he received a B.S. in Forestry at the Chinese Cultural University in Taipei, Taiwan. He continued his studies at Griffith University in Queensland, Australia, receiving a M.S. degree with honors in July 2004, completing his thesis, “Isolation and Identification of Microorganisms Associated with the Cyanobacterium Cylindrospermosis raskiborsii”. Upon fulfillment of his M.S. degree, JayHow gained experience isolating and characterizing chlorophyll pigments and inner and outer membrane components in chloroplasts, working under the supervision of Dr. Chi-Ming Yang at the Research Center for Biodiversity at the Academia Sinica, Taiwan.
|
JayHow’s Ph.D. studies focus on the isolation and purification of the amino acid transporter OEP16 from Pisum sativum (pea plant) and Spinacia oleracea (spinach). OEP16 is a small, 16 kDa membrane-integral amino acid transporter protein for which the structure has yet to be revealed. JayHow uses analytical centrifugation (sucrose gradients) to isolate the inner and outer membrane constituents of the pea plant and is able to identify OEP16 through the use of SDS-PAGE and immunoblotting techniques. Following purification, crystals of OEP16 have been produced. Further X-ray diffraction studies will be attempted in order to solve the structure of OEP16, which would contribute greatly to the field of membrane transport proteins, notoriously challenging to purify and characterize.
Another focus of JayHow’s research is the isolation, purification, and subsequent crystallization of the intact ATP synthase from spinach. ATP synthase is an enzyme responsible for producing ATP from ADP and inorganic phosphate using the energy obtained from an electrochemical gradient occurring across the membrane separating the lumen and the stroma in the chloroplast. ATP synthase has a prominent role in the photosynthetic electron transport chain. Currently, no structure of the intact ATP synthase exists. Elucidating the structure of the intact ATP synthase would be of importance for the bioenergy and photosynthesis community.
|
|
| | |
















