Research of the group of Petra Fromme
at Arizona State University
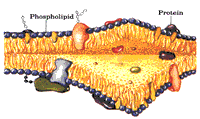
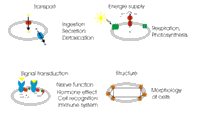
The research in our group focuses on the structural biochemistry and biophysics of membrane proteins. Membrane proteins perform most of the important processes in all living cells. For example, respiration, photosynthesis, cell communication, cell import/export, cell growth and recognition are catalyzed and regulated by membrane proteins. These proteins do not act in an isolated way; they rather perform communication within the cell by binding and releasing of cofactors and soluble signal-transducing proteins. Membrane proteins are also key player in infectious diseases as they mediate entry of viral and bacterial pathogens into the host cell and also play an important role in the cell defense against the pathogens. The main step for the elucidation of the complex in whole living cells is the understanding of the structure, dynamics and function of the membrane proteins that play the key role in these processes. Our research field is of a very interdisciplinary nature and includes biochemical investigations, molecular biology, spectroscopy, crystallization, X-ray structure analysis, as well as theoretical investigations. Petra Fromme's group is part of a large international collaboration who are pioneers the new field of serial femtosecond nanocrystallography using Free electron lasers, where structure determination is based on femtosecond X-ray diffraction from a stream of nanocrystals, which will allow the determination of molecular movies of biomolecules at work in the future. The Fromme group has two major biological fields of interest: Photosynthesis and Infectious Diseases. Photosynthesis is the main process on earth converting light energy provided by the sun into chemical energy. It is the unique energy source for all higher life on earth and produces all the oxygen in the atmosphere. The work on Photosynthesis includes the investigation of the structure and function of the large membrane protein complexes involved in the primary processes of photosynthesis and the development of an artificial oxygen evolving complex in DNA nanocages, and is part of the ASU Center for Bio-Inspired Solar Fuel production. A special focus for our studies is the structure and function of the large bio-solar energy converters, Photosystem I, Photosystem II, and the ATP-Synthase, an enzyme that functions as a molecular motor. We also use time-resolved femtosecond nanocrystallography to determine a molecular movie of water splitting. Another exciting project deals with transport processes across membranes, with a special focus on transport into cell organelles. The work on important viral, bacterial and human membrane proteins is the focus of the ASU Center for Membrane Proteins in Infectious Diseases led by Petra Fromme. The center involves a collaboration of 11 groups at ASU. We have selected 50 target proteins, which play important roles in the infection cycle. The work involves new method development for expression, purification, biophysical characterization, crystallization and structure determination of the membrane proteins. Overview The unraveling of the structure and function of membrane proteins is one of the most challenging goals in the post-genomic era.
The main step for the elucidation of these complex processes in whole living cells is the understanding of the structure, dynamics and function of the membrane proteins, which play the key role in these processes. The great importance of membrane proteins as the major key player in the function of all living cells, being targets for more than 50% of all drugs, stands in contrast to the lack of knowledge on their structure and function, the main deficit being the lack of structural information. More than 25 000 structures of soluble proteins have been solved so far by X-ray structure analysis or NMR spectroscopy, whereas only structures of less than 50 functional and structural different membrane proteins have been worldwide solved so far. The crystallization is the rate-limiting step for the elucidation of their structure and function. The deeper understanding of the physical chemical processes, which occur during membrane protein crystallization, is therefore one major research topic of my group.
The main step for the elucidation of these complex processes in whole living cells is the understanding of the whole pictures of the structure, dynamics and function of the membrane proteins as the key players in important cell processes. Spectroscopic spectator labels will be introduced to allow the investigation of these processes in reconstituted protein, the protein detergent micelles and even in the single crystals. These investigations are of a very interdisciplinary nature and include biochemical investigations, molecular biology, spectroscopy, structural investigations by X-ray structure analysis as well as theoretical investigations. Current Research Projects
Dr. Fromme directs the Center for Applied Structural Discovery (CASD) at Arizona State University’s Biodesign Institute, which unites a team of complementary research professionals from a wide range of disciplines, including biology, chemistry, physics and engineering, to develop and apply groundbreaking technologies and methodologies to accelerate the rate at which we discover the structure and associated function of biomolecules. Breakthrough knowledge will accelerate progress on the path to technical innovations that improve human health and provide plentiful clean energy and food for future generations. (https://biodesign.asu.edu/applied-structural-discovery)
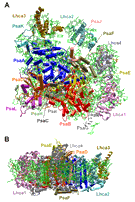
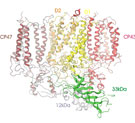
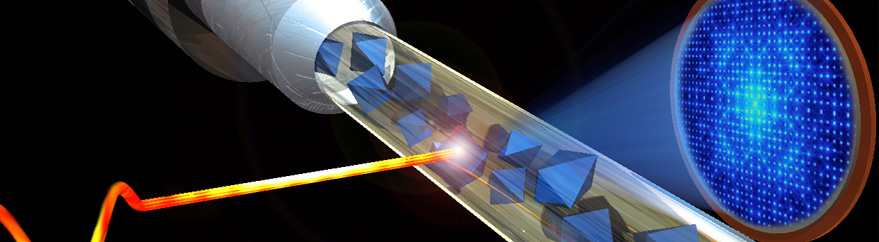
Femtosecond pulses of X-ray free electron lasers are used to investigate the structure of membrane proteins. The project aims to develop the method of femtosecond crystallography for the structure determination of membrane proteins, where X-ray structure analysis is based on hundreds of thousands of X-ray diffraction patterns from a stream of fully hydrated nano/microcrystals of membrane protein crystals. The determination of membrane proteins structures solved to date often involves a long process where it took years (or sometimes even decades) to grow large, well-ordered crystals suitable for X-ray structure determination. Furthermore, X-ray damage is a major problem for many membrane protein crystals, especially when they contain redox active cofactors, and it also imposes a limitation for X-ray diffraction on microcrystals, even under cryogenic conditions. This project aims to open an exciting new avenue for membrane protein crystallography, where hundreds of thousands of diffraction patterns can be collected in a few minutes from fully hydrated micro/nano crystals in their native conditions at room temperature, using X-ray laser pulses that are so short that no X-ray damage occurs during data collection. This method will be used to solve novel X-ray structures of membrane proteins.
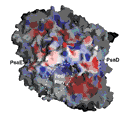
Membrane proteins represent > 60 percent of all drug targets and are also key players in the pathogenesis of infectious diseases. A critical step for the elucidation of the complex processes that are catalyzed by membrane proteins is an understanding of the structure, dynamics and function of membrane proteins. Our knowledge of processes catalyzed by membrane proteins suffers mainly from the lack of information concerning their structure as less than 300 different membrane protein structures are known at present. The MPID Center targets membrane proteins of important viral and bacterial pathogens, their infectious pathways, and molecules involved in host defense against the pathogens. The structural determination of each of the targets may provide important clues for the understanding of the infectious disease pathways and can therefore form the basis for the treatment and prevention of infectious diseases. (http://mpid.asu.edu/)
Completed Projects
|
||||||||||||||||||||||||||||
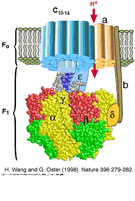
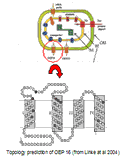
 The X-ray structure of membrane proteins provides detailed structural information on one static functional state of the protein and is therefore the basis for the understanding of their function. It is exciting to extend this picture to the understanding of more complex processes essential to the function of the membrane proteins, e.g., the interaction with signaling molecules, proteins, cofactors, and drugs. Therefore, a further challenge in my work on membrane proteins also lies in the unraveling of the dynamics of membrane proteins.
The X-ray structure of membrane proteins provides detailed structural information on one static functional state of the protein and is therefore the basis for the understanding of their function. It is exciting to extend this picture to the understanding of more complex processes essential to the function of the membrane proteins, e.g., the interaction with signaling molecules, proteins, cofactors, and drugs. Therefore, a further challenge in my work on membrane proteins also lies in the unraveling of the dynamics of membrane proteins. 
 MPID is a PSI:Biology center investigating the structures of viral, bacterial and human membrane proteins involved in pathogenesis. Under the leadership of the principal investigator, Petra Fromme, the MPID Center is trying to use the biological theme of membrane proteins in infectious diseases as the basis for the determination of novel membrane protein structures and to develop new technologies for high throughput membrane protein expression, isolation, functional characterization, crystallization and structure determination.
MPID is a PSI:Biology center investigating the structures of viral, bacterial and human membrane proteins involved in pathogenesis. Under the leadership of the principal investigator, Petra Fromme, the MPID Center is trying to use the biological theme of membrane proteins in infectious diseases as the basis for the determination of novel membrane protein structures and to develop new technologies for high throughput membrane protein expression, isolation, functional characterization, crystallization and structure determination. BioXFEL is a Science and Technology center using X-ray lasers to image biomolecular machines in motion. X-ray lasers provide snapshots of unprecedented clarity depicting the building blocks of life. The motions and arrangements of these blocks underlie all biological function. The mission of BioXFEL is:
BioXFEL is a Science and Technology center using X-ray lasers to image biomolecular machines in motion. X-ray lasers provide snapshots of unprecedented clarity depicting the building blocks of life. The motions and arrangements of these blocks underlie all biological function. The mission of BioXFEL is: