Research
Not exhaustive list of research topics, please constact us directly for the most up-to-date list of research opportunities.DNA and RNA biophysics and nanotechnology: theory and experiments
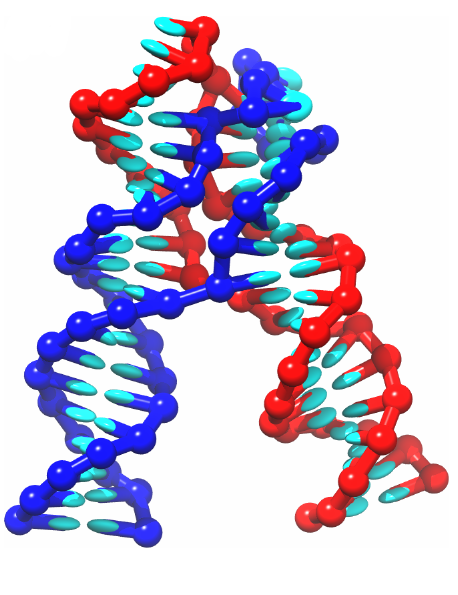
We work on developing and applying coarse-grained models of nucleic acids. Our DNA model, oxDNA, is parametrized to reproduce quantitatively structural, mechanical and thermodynamic properties of DNA. It has been successfully used to study both fundamental physics of DNA molecules as well as DNA nanotechnological systems. DNA nanotechnology is a rapidly developing field which uses DNA molecules as basic building blocks to assemble nanoscale structures as well as active molecular devices. Computer simulations using our coarse-grained model provide insight into the functioning of such systems and give us better understanding of underlying physical processes. Besides theoretical modeling, we also carry out experiments that are guided by the computer simulations using our software. Our current DNA/RNA nanotechnology projects comprise: 1) Tool development (new models and visualization and analysis tools); 2) Application to new biomaterial development, combining multiscale simulation with experimental studies; 3) Biotechnology application: Development of new nanostructures for biomedical applications.The code implementing our model, oxDNA, is available as a free software. The code website also provides examples and tutorials.
Principles of self-assembly
How to design building blocks so that they reliable assemble into a target structure? Biology can achieve structure of extraordinary complexity by self-assembly of different building block, something that nanotechnology has been trying to mimic for a long time. We employ methods of computer simulations and statistical physics to answer questions such as "What is the minimal number of blocks needed to achieve a target structure", and "How to avoid possible kinetic traps and undesired assemblies?"Machine learning for sequence ensembles
Aptamers are short DNA or RNA molecules selected by in-vitro protocol (SELEX) to bind to a certain molecular target (protein, small molecule, specific cell surface etc.) with applications for diagnostics and therapeutics. We use machine learning to study the sequence ensemles, identify functional motifs and generate in-silico new binders.Immunostimulatory motifs in RNA
We employ machine learning and statistical physics methods to quantify immunostimulatory motifs in viral genomes, as well as in cells, with applications to diagnostics, treatment as well as bionanotechnology.Smartgrid: Control of reactive power from photovoltaic generators
Smartgrid refers to power grid which incoporates modern computer communication technology to improve efficiency and reliability of production, distribution and consumption of electricity. One of the big challenges that the power grid faces at the moment is the integration of renewable sources of energy.Together with Kostya Turitsyn, Misha Chertkov and Scott Backhaus from Los Alamos National Laboratory, we investigated ([1], [2], [3]) the possibilities of controlling reactive power flow in a distribution network that contains photovoltaic generators (typical example would be an urban neighborhood with houses with photovoltaic cells on roofs). We considered both global and local control of the generated reactive power and found optimization schemes that can reduce thermal losses in the distribution line and improve voltage stability.